Your gateway to endless inspiration
Microbiology - Blog Posts

Daily Doodles- Day 190- 22/10/24
nature pattern hand! I plan to make this into a sticker soon! (not this drawing specifically, but the idea)
The tag for this is #agdoodles
Cupid Bacteria anyone?







bacteria sooooooooo cute (google search emoji kitchen)

Curious about how to send research to the International Space Station or how to get involved with NASA missions as a college student? Ask our experts!
Through our Student Payload Opportunity with Citizen Science, or SPOCS, we’re funding five college teams to build experiments for the International Space Station. The students are currently building their experiments focusing on bacteria resistance or sustainability research. Soon, these experiments will head to space on a SpaceX cargo launch! University of Idaho SPOCS team lead Hannah Johnson and NASA STEM on Station activity manager Becky Kamas will be taking your questions in an Answer Time session on Thurs., June 3, from 12-1 p.m. EDT here on our Tumblr! Make sure to ask your question now by visiting http://nasa.tumblr.com/ask. Hannah Johnson recently graduated from the University of Idaho with a Bachelor of Science in Chemical Engineering. She is the team lead for the university’s SPOCS team, Vandal Voyagers I, designing an experiment to test bacteria-resistant polymers in microgravity. Becky Kamas is the activity manager for STEM on Station at our Johnson Space Center in Houston. She helps connect students and educators to the International Space Station through a variety of opportunities, similar to the ones that sparked her interest in working for NASA when she was a high school student. Student Payload Opportunity with Citizen Science Fun Facts:
Our scientists and engineers work with SPOCS students as mentors, and mission managers from Nanoracks help them prepare their experiments for operation aboard the space station.
The Vandal Voyagers I team has nine student members, six of whom just graduated from the Department of Chemical and Biological Engineering. Designing the experiment served as a senior capstone project.
The experiment tests polymer coatings on an aluminum 6061 substrate used for handles on the space station. These handles are used every day by astronauts to move throughout the space station and to hold themselves in place with their feet while they work.
The University of Idaho’s SPOCS project website includes regular project updates showing the process they followed while designing and testing the experiment.
Make sure to follow us on Tumblr for your regular dose of space: http://nasa.tumblr.com.
The As, Gs, Cs and Ts of the Space Station: First In-Space Microbe Identification
Being able to identify microbes in real-time aboard the International Space Station, without having to send them back to Earth for identification first, would be totally amazing for the world of microbiology and space exploration.

The Genes in Space 3 team turned that possibility into a reality this year, when it completed the first-ever sample-to-sequence process entirely aboard the space station.
The ability to identify microbes in space could aid in the ability to diagnose and treat astronauts in real time, as well as assisting in the identification of life on other planets. It could also benefit other experiments aboard the space station.
HELPFUL SCIENCE HINT: Identifying microbes involves isolating the DNA of samples, and then amplifying – or making lots and lots (and LOTS) of copies - of that DNA that can then be sequenced, or identified.
As part of regular monitoring, petri plates were touched to various surfaces of the space station. NASA astronaut Peggy Whitson transferred cells from growing bacterial colonies on those plates into miniature test tubes, something that had never been done before in space (first OMG moment!).

Once the cells were successfully collected, it was time to isolate the DNA and prepare it for sequencing, enabling the identification of the unknown organisms – another first for space microbiology.
Enter Hurricane Harvey. *thunder booms*

“We started hearing the reports of Hurricane Harvey the week in between Peggy performing the first part of collecting the sample and gearing up for the actual sequencing,” said Sarah Wallace, the project’s primary investigator.
When our Johnson Space Center (JSC) in Houston became inaccessible due hurricane conditions, Marshall Space Flight Center’s Payload Operations Integration Center in Huntsville, Alabama worked to connect Wallace to Whitson using Wallace’s personal cell phone.
With a hurricane wreaking havoc outside, Wallace and Whitson set out to make history.

The data were downlinked to the team in Houston for analysis and identification.
“Once we actually got the data on the ground we were able to turn it around and start analyzing it,” said Aaron Burton, the project’s co-investigator. “You get all these squiggle plots and you have to turn that into As, Gs, Cs and Ts.”
Those As, Gs, Cs and Ts are more than just a nerdy alphabet – they are Adenine, Guanine, Cytosine and Thymine – the four bases that make up each strand of DNA and can tell you what organism the strand of DNA came from.

“Right away, we saw one microorganism pop up, and then a second one, and they were things that we find all the time on the space station,” said Wallace. “The validation of these results would be when we got the sample back to test on Earth.”
Soon after, the samples returned to Earth aboard the Soyuz spacecraft, along with Whitson.
With the samples now in the team’s JSC lab, tests were completed in ground labs to confirm the findings from the space station. They ran the tests again and again, and then once more, to confirm accuracy. Each time, the results were exactly the same on the ground as in orbit. (second OMG moment!)

“We did it. Everything worked perfectly,” said Sarah Stahl, microbiologist.
This capability could change future space exploration.
“As a microbiologist,” said Wallace, “My goal is really so that when we go and we move beyond ISS and we’re headed towards Mars or the moon or wherever we are headed to, we have a process that the crew can have that great understanding of the environment, based on molecular technology.”
For more information, follow @ISS_Research.
Make sure to follow us on Tumblr for your regular dose of space: http://nasa.tumblr.com.







So far, at 50. Lol.
i complain alot when it comes to uni and my course, but not gonna lie, here on my final year i've started to fall in love with it again, the way the fascination started when i was younger and learning new things was exciting.
throughout learning it always felt like i was not built for it, that I just cannot for the life of me focus and dedicate myself on anything. and i was just doubting myself and i should change courses or drop out because I was not meant to do this. and now on my second last semester, things kinda clicked. It may be hard for me to understand and learn, but it's worth it. To see the universe in all of its beauty, its ugliness, its complexity, its charm; it's a struggle but I'll endure it for you.
and I find myself really hoping I get to continue down in the stream of sciences and contribute to something for nature and for humanity as well, or at least deepen my understanding of how this universe works and widen my view of how intricate and special this world we live in actually is, how caring it is, how every single thing is worth something, and nothing from nature is ever truly useless
![[Mitosis In Garlic Roots]](https://64.media.tumblr.com/8f107e97d6549ff315eb6aff14b4137e/tumblr_ohvmnvR8NM1vj7kdmo1_500.jpg)
![[Mitosis In Garlic Roots]](https://64.media.tumblr.com/adbd5e46f3bb41ed49c870158b591cbe/tumblr_ohvmnvR8NM1vj7kdmo2_500.jpg)
![[Mitosis In Garlic Roots]](https://64.media.tumblr.com/1e2b58752ded37c7d73d65f71b0b5ede/tumblr_ohvmnvR8NM1vj7kdmo3_500.jpg)
![[Mitosis In Garlic Roots]](https://64.media.tumblr.com/29d207c139caf42101f9a9454a18507b/tumblr_ohvmnvR8NM1vj7kdmo4_500.jpg)
[Mitosis in garlic roots]
Crash Course (Mitosis; Splitting Up Is Complicated)

Fungal tissues – the fungal mantle around the root tip and the fungal network of tendrils that penetrates the root of plants, or Hartig Net, between Pinus sylvestris plant root cells – in green. Ectomycorrhizal (ECM) fungi help trees tolerate drought and boost the productivity of bioenergy feedstock trees, including poplar and willow.
Via Berkeley Lab: The sclerotia are in the soil!
More: How Fungi Help Trees Tolerate Drought (Joint Genome Institute)
One thing I absolutely adore about research papers is when they’re named something really sharp and professional like ‘Pseudomonas sp. Strain 273 Incorporates Organofluorine into the Lipid Bilayer during Growth with Fluorinated Alkanes‘ and then they put a graphical abstract that looks like this

Got some microorganisms that are soon to be examined 🐚🐟🔬





Tardigrades: ‘Water bears’ stuck on the moon after crash
The moon might now be home to thousands of planet Earth’s most indestructible animals.
Tardigrades - often called water bears - are creatures under a millimetre long that can survive being heated to 150C and frozen to almost absolute zero.
They were travelling on an Israeli spacecraft that crash-landed on the moon in April.
And the co-founder of the organisation that put them there thinks they’re almost definitely still alive.
The water bears had been dehydrated to place them in suspended animation and then encased in artificial amber.
“We believe the chances of survival for the tardigrades… are extremely high.”






Journey to the Microcosmos- Flatworms: Simple Wiggly Tubes
Images Originally Captured by Jam’s Germs
Quote Voiced by Jam’s Germs

The bacteria wars are coming. Researchers at Tel Aviv University have pitted “good” bacteria against “bad” bacteria and the good guys, it appears, are winning.
If the system can be scaled, this new approach could potentially replace antibiotics, which are increasingly struggling against antibiotic-resistant “superbugs.” For the TAU study, the researchers used a toxin injection system known as a “Type 6 Secretion System.” It’s usually deployed by pathogenic (“bad”) bacteria. They introduced the system into a “friendly” bacterium, Vibrio natriegens, which is not harmful to humans. The researchers described their technology as similar to a microscopic poison arrow shot from a good bacterium to eliminate a bad bacterium under specific conditions. “The system that we built allows us to engineer ‘good’ bacteria that can recognize pathogenic bacteria, attack them with toxins, and neutralize them,” explains Dr. Dor Salomon, who co-led the study. “We know how to change and control every component in the system and create a bacterium that neutralizes different strains of bacteria. This is proof of feasibility, showing that we have the knowledge and ability to create bacteria that take advantage of this killing system and may serve as antibiotic treatments. ”The current bacteria prototype is best suited for bugs that occur naturally in saltwater. This is a growing concern, as fish and seafood constitute a major food source in many regions of the world. “Their productivity is severely impaired as a result of bacteria-borne diseases,” Solomon notes, “and since we want to avoid pouring antibiotics into aquaculture farms, a biological solution such as the one we have developed is an effective alternative.” The system will eventually be adapted to treat pathogenic bacteria in humans, farm animals and plants. Tel Aviv University has filed a patent application through Ramot, the university’s technology-transfer company. In addition to Solomon, Dr. Biswanath Jana and Kinga Kappel of the department of clinical microbiology and immunology at TAU’s Sackler Faculty of Medicine participated in the research. The results were published this month in the scientific journal EMBO Reports.





"Pelomyxa are giant amoeba, capable of growing as much as 5 millimeters in length. So it doesn't seem like they should be that difficult to find. In fact, Jame--our master of microscopes--found hundreds of them filling up his pond tank, completely visible without the help of a microscope. They were so large that he actually took one and touched it. So, if you would like to know what it feels like to pet an amoeba, he is now an expert. He says, "It's squishy.""
Journey to the Microcosmos- The Microbe That's Big Enough to Pet
Images Originally Captured by Jam's Germs
Quote Voiced by Hank Green


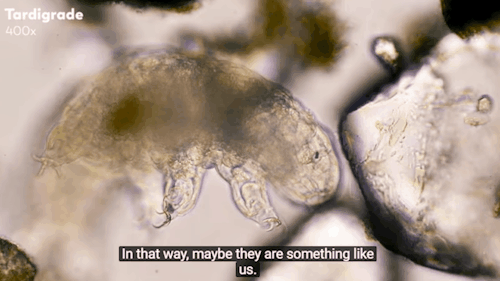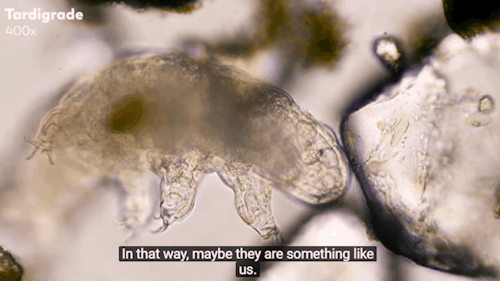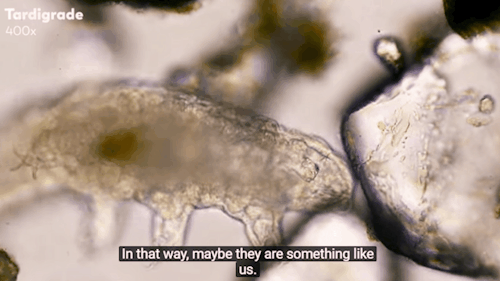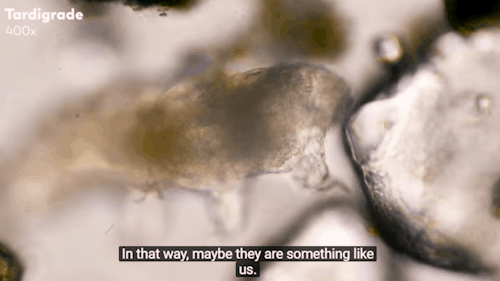


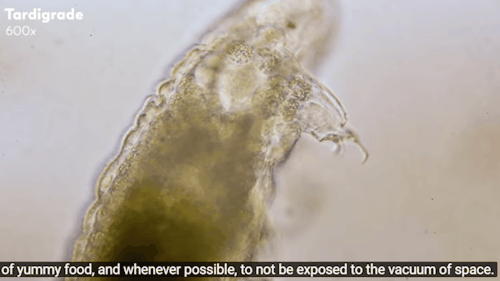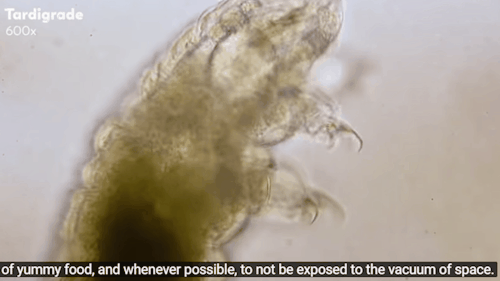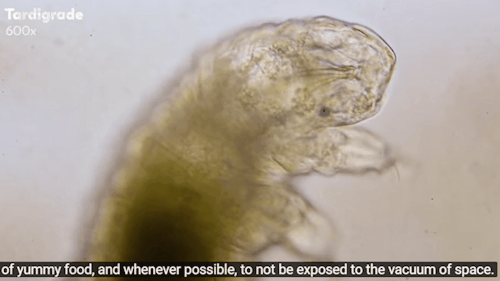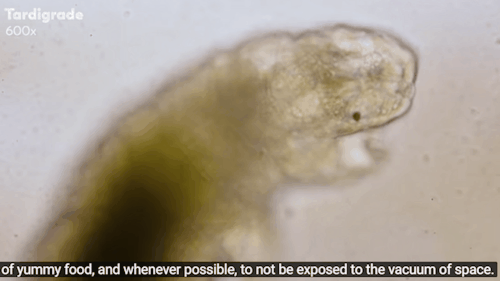
Journey to the Microcosmos: Tardigrades: Chubby, Misunderstood, & Not Immortal
Images originally captured by Jam’s Germs
Thank you @airyearthgirl for inspiring me to gif these amazing lines

